Introducing Nucleotide Transformer v3 (NTv3) for Genomics
NTv3 revolutionizes genomic prediction and design with its multi-species foundation model.
Understanding NTv3 in Genomics
Genomic prediction and design now require models that connect local motifs with megabase scale regulatory context across various organisms. Nucleotide Transformer v3 (NTv3) is InstaDeep’s innovative multi-species genomics foundation model tailored for this environment. It unifies representation learning, functional track and genome annotation prediction, and controllable sequence generation in a single framework designed to process 1 Mb contexts at single nucleotide resolution.
Advancements Over Previous Models
Earlier Nucleotide Transformer models demonstrated strong features for molecular phenotype prediction through self-supervised pretraining on thousands of genomes. The original series included models ranging from 50M to 2.5B parameters, trained on 3,200 human genomes and 850 additional genomes from diverse species. NTv3 retains this sequence-only pretraining concept while extending it to longer contexts and incorporating explicit functional supervision along with a generative mode.
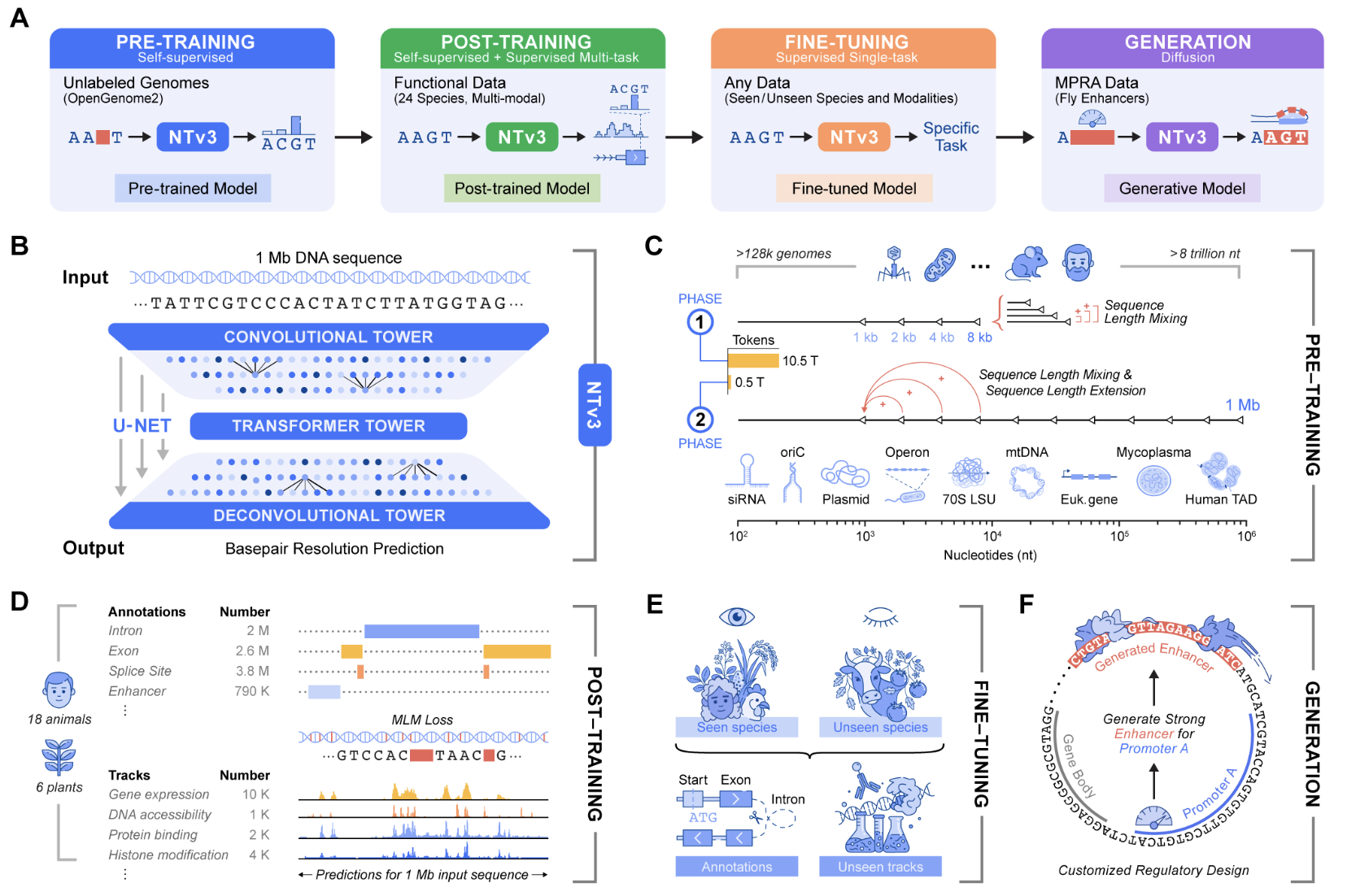
Architecture for 1 Mb Genomic Windows
NTv3 employs a U-Net style architecture targeting extended genomic windows. A convolutional downsampling tower compresses the input sequence, a transformer stack models long-range dependencies in that compressed space, and a deconvolution tower restores base-level resolution for prediction and generation. Inputs are character-tokenized over A, T, C, G, N with special tokens like <unk>, <pad>, <mask>, <cls>, <eos>, and <bos>. Each sequence length must be a multiple of 128 tokens, with padding used to enforce this.
The smallest public model, NTv3 8M pre, comprises approximately 7.69M parameters with a hidden dimension of 256 and 2 transformer layers. Conversely, NTv3 650M features a hidden dimension of 1,536, 12 transformer layers, and species-specific conditioning layers for prediction heads.
Extensive Training Data
The NTv3 model is pretrained on 9 trillion base pairs from the OpenGenome2 resource through base resolution masked language modeling. This phase is followed by post-training with a joint objective integrating self-supervised learning and supervised learning on around 16,000 functional tracks and annotation labels from 24 species.
Performance Metrics and Benchmarking
Post-training, NTv3 achieves state-of-the-art accuracy for functional track prediction and genome annotation across species. It surpasses significant sequence-to-function models and previous genomics foundation models on public benchmarks, along with the new Ntv3 Benchmark—defined as a controlled downstream fine-tuning suite with standardized 32 kb input windows and base resolution outputs. The Benchmark encompasses 106 tasks covering long-range, single nucleotide, cross-assay, and cross-species challenges.
From Prediction to Controllable Sequence Generation
Beyond prediction abilities, NTv3 can be fine-tuned into a controllable generative model utilizing masked diffusion language modeling. This capability allows the model to receive conditioning signals encoding desired enhancer activity levels and promoter selectivity, filling masked segments of DNA in alignment with specified conditions.
The team has successfully designed 1,000 enhancer sequences with predefined activity values, which were validated in vitro using STARR seq assays in collaboration with the Stark Lab. Results indicate these generated enhancers maintain the intended ordering of activity levels, achieving more than 2-times improved promoter specificity relative to baselines.
Key Takeaways
- Comprehensive Model for Genomics: NTv3 integrates representation learning, functional track prediction, genome annotation, and controllable sequence generation within a U-Net style architecture for 1 Mb nucleotide resolution across 24 species.
- Extensive Training Data: Utilizing 9 trillion base pairs with a joint objective expands the model's applications and accuracy.
- State-of-the-Art Performance: Surpassing previous models on public benchmarks, NTv3 proves its efficacy on the Ntv3 Benchmark with 106 standardized long-range tasks.
- Innovative Enhancer Design: The backbone model enables the generation of validated enhancer sequences, showcasing improved specificity and activity levels.
Сменить язык
Читать эту статью на русском